
|

|

|
BioTapestry Web Client Quick Start |
||

|

|

|
BioTapestry Web Client Quick Start |
||
Contents
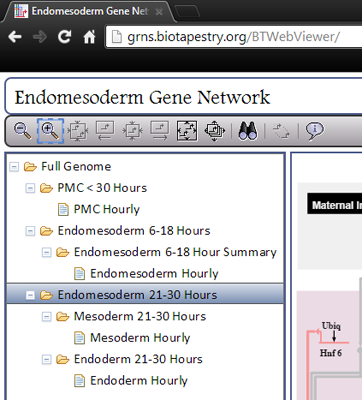
BioTapestry organizes a network model in a hierarchical series of models and sub-models, and displays this hierarchy in a tree view on the left side of the browser window. You can choose the model you want to view by clicking on it in the tree view.
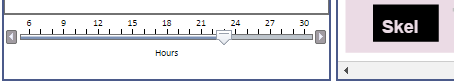
Whenever an "hourly" view is selected (the lowest level in the hierarchy), the viewer displays a time slider in the lower left corner that allows you to select the hour you wish to view by dragging the slider. Clicking the arrow 'bumper' to the left or right of the slider also moves it one increment per click.
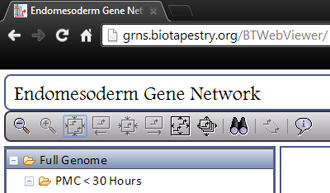
The BioTapestry Viewer toolbar provides several important functions. As shown in the picture at left, there are buttons for (going left to right) zooming out one step, zooming in one step, zooming to all selected items (only active if something is selected), cycling backwards through the selected items (only active if multiple items are selected), zooming to show the current selected item (only active if multiple items are selected), cycling forwards through the selected items (only active if multiple items are selected), zooming to view the current model, zooming to bound all the models in the hierarchy, searching for genes/nodes, clearing current selections (only active if something is selected), and viewing the about dialog.
To select a gene, node, or link segment, just click on it. For genes, make sure to click in the areas highlighted in orange shown in the picture at left: either the gene name or the gene itself. To select more than one thing, hold down the Shift key while clicking on each new item. Use the Clear Current Selections toolbar button to clear all current selections, or, click on an empty section of the model.
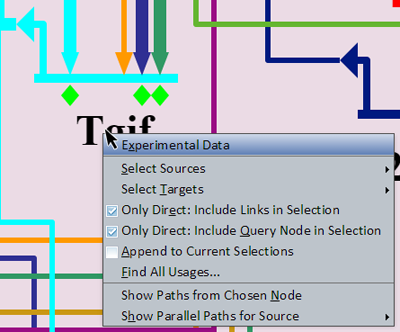
Right-clicking on a gene brings up a menu of options for that gene (see picture at left). (Note to Mac users: If you're using a one-button mouse, press the Control key while clicking the mouse to do this.) Make sure to click in the areas highlighted in orange shown in the picture in the previous section: either the gene name or the gene itself. (If you miss these areas, you will instead activate the menu for the underlying region or an inbound link.) Also, trying to select genes that are grayed out with no active inputs or outputs will not bring up this menu. Possible operations are:
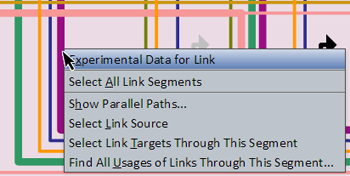
Right-clicking on a link segment brings up a menu of options for that link (see picture at left). (Note to Mac users: If you're using a one-button mouse, press the Control key while clicking the mouse to do this.) Possible operations are:
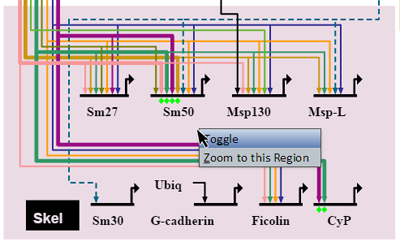
Right-clicking on a colored region brings up a menu of options for that region (see picture at left). (Note to Mac users: If you're using a one-button mouse, press the Control key while clicking the mouse to do this.) Possible operations are:
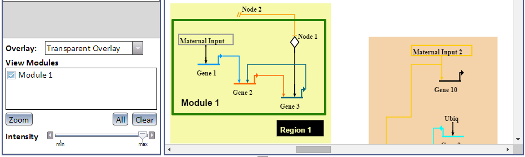
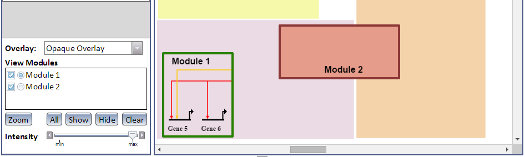
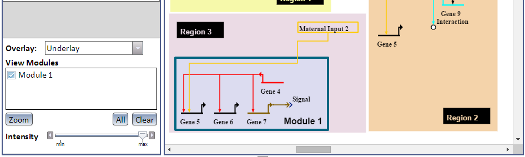
Overlays are a visual aid which allow you to draw rectangular boxes (called Network Modules) around nodes and links in a model. Different models may use different types of Overlays, or none at all, and a single Overlay can contain multiple Network Modules. Overlays can be one of three visual types, which effects how their Network Modules are displayed (see left image):
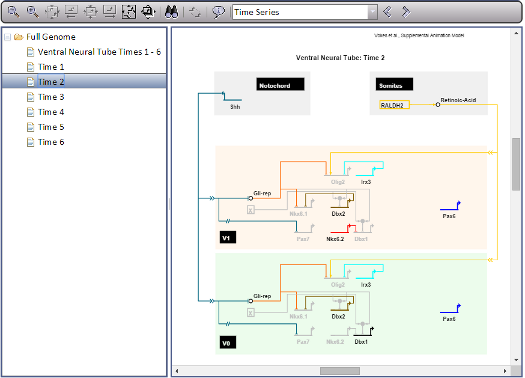
User Paths allow you to create a 'path' stepping forwards and backwards through different levels and models in your network model. They are useful for showing a specific route through your network model, and can incorporate Overlay settings and use specific time point settings in models which have them. If a model has User Paths defined, a new drop-down and arrow buttons will be visible on the toolbar to the right of the About button (see image at left). To disable any current User Path, set the drop-down to 'None'. While the drop-down is set to 'None', the forward and back arrow buttons will be disabled. Clicking on either button will advance you one step forwards or backwards along the path.
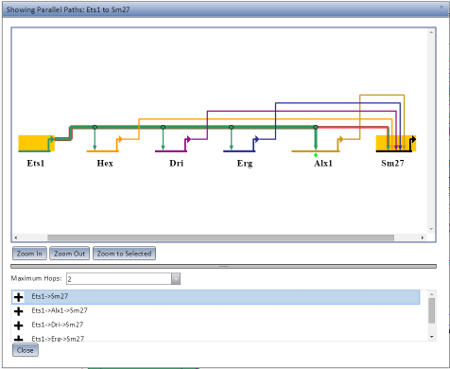
The Pathing Display opens when you choose Show Parallel Paths from Source or Show Parallel Paths from Chosen Node from the Gene Information pop-up. It will preferentially open as a new browser tab or window if it can (depending on your browser settings), but if not it will open as a dialog in the current browser window. An example of the Pathing Display open as a dialog is visible at the left.
The pathing model displayed will show all nodes along the possible route, with the current route highlighted in selection colors. There are three zoom options (Zoom In, Zoom Out, and Zoom to Selected) which behave the same as in the main application display. The maximum number of hops to allow for a potential route can be set using the Maximum Hops drop-down. A given rooute can be highlighted by selecting it from the list below the Maximum Hops limit.
Footnotes describing features of the network diagram are shown as red numbers. If you move your mouse over the footnote, the associated text will be displayed in the box at the bottom of the BioTapestry window (see picture at left). Clicking on the note will keep the text displayed even if you move your mouse elsewhere.
Last updated: January 14, 2015